ELI15: PCR and PCR Testing
The science behind molecular COVID tests
Posted by ekr on 14 Sep 2022
As pretty much everyone is now aware, there are two main kinds of COVID test:
- At-home based antigen tests (often called "lateral flow")
- Lab-based molecular tests (often called "PCR" [though not all molecular tests are PCR—2022-09-14])
Lateral flow and PCR are both descriptions of the technology used in the test, but unless you already know what they are, they're just tech jargon. The purpose of this post is to explain how PCR works at the "explain it like I'm fifteen" level. To that end, I'll be omitting most of the chemistry and focusing on the main clever ideas, with some external cites for those who want to learn more.
Background: DNA #
As you no doubt know Deoxyribonucleic acid (DNA)—this is the last time you will need to read "deoxyribo..."—carries the genetic code that directs the development of humans and most (but not all, as we'll see) living things. The basic structure of DNA is of a sequence of small molecular subunits (nucleotides). Nucleotides generally have the same basic structure, which consists of a common "backbone" consisting of a sugar, a phosphate group, plus another chemical group called a nucleobase:
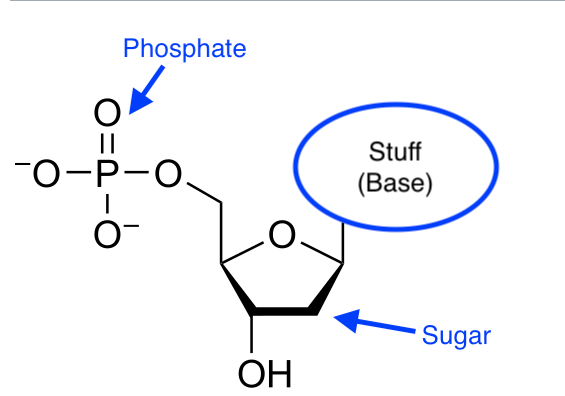
[Modified version of a diagram from Wikipedia]
Each type of nucleotide has a different nucleobase.
You can build a chain of nucleotides by attaching the sugar group of nucleotide 1 to the phosphate group of nucleotide 2 and then the sugar group of nucleotide 2 to the phosphate group of nucleotide 3, and so on. This is true no matter what nucleobases are attached to the backbone. There are four main bases, adenine, cytosine, guanine, and thymine, which gives us a 4-ary code (unlike the binary code used by computers). It's canonical to refer to these by their leading letters: A, C, G, T.
Instead of being a single chain, normally DNA exists as a pair of chains (with each chain often being called a strand). The key thing to know is that not any two strands can hook up. Instead, the base pairs are complementary: with adenine pairing with thymine and guanine pairing with cytosine, like this:
[Image from Wikipedia by Madeleine Price Ball.]
Thus, the sequence of bases on the first strand determines the sequence of bases on its paired strand. This means that most of what you need to know about a given DNA molecule is encoded in the sequence of bases. It's this sequence which determines the DNA code for an organism.[1]
If you take two pieces of DNA with complementary sequences, they will tend to hybridize with each other to form a pair of strands. This will also work—though not quite as well—if the sequences are close but not identical, which can be used to measure "closeness" of two sequences; this was a more useful technique before sequencing became fast and cheap.
You don't need to know this to understand PCR, but the actual DNA molecule is arranged in a characteristic double helix structure, which looks like this:
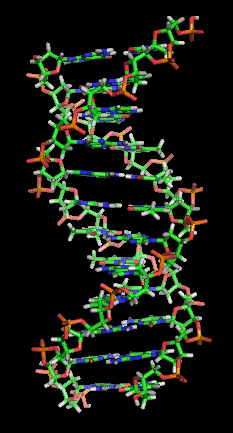
[Image from Zephyris via Wikipedia]
DNA Replication #
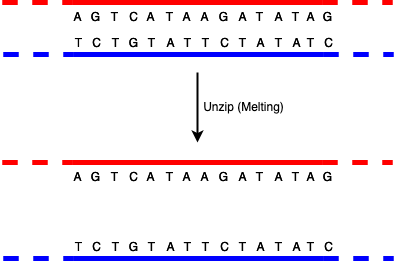
The way that DNA replicates—for instance when a cell divides into two, requiring two copies of the DNA, one for each cell— is that the paired strands unzip to form two single strands (this is called "melting").
Note that in this diagram I'm only showing short strands with a small number of bases. The dashed regions are intended to indicate that the DNA strand just goes on indefinitely, but I'm not going to show it.
Next, an enzyme called DNA polymerase builds another paired strand onto each strand using the complementary bases in the ambient cellular environment. The result is now two DNA double helices, each of which is (hopefully)[2] an exact copy of the first one, and each of which contains one of the original strands and one newly created strand, which used the original strand as the template.
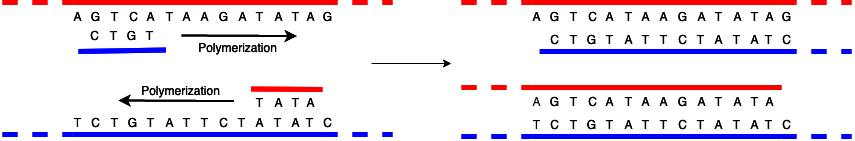
The diagram below shows the process of replicating the unzipped DNA.
There are several more important things to notice here. First, the two strands are built in opposite directions. I've colored things red and blue to help keep track, with the blue chain polymerizing left to right (built off the red template which goes the other way) and the red right to left (built off the blue template, which goes the other way). Note that in reality both strands have the same chemistry, it's just that the backbones are facing opposite directions, and of course the bases are complementary.
Second, the process gets kicked off by having a primer, which is a short piece of DNA that is (of course) complementary to the strand being replicated. Because polymerization only happens in one direction, however, the primer ends up being one end of the replicated DNA strand, with everything on the other side of the primer just not being replicated. You can see this in the diagram above, there the replicated blue strand has nothing on the left of the CTGT (losing the T that was there in the original, as well as everything else to the left which I didn't show) and the replicated red strand has nothing on the right of the TATA, losing the C (as well as everything else to the right). Note that the top red and bottom blue strands are actually the originals, and so extend in both directions.
In normal DNA replication in the body, you'd want to replicate the entire strand and so the primer would be attached to the end of the chain (there's some special biochemistry for this that we don't need to go into), but in PCR, the primers are just little snippets of single-stranded DNA that get attached to the DNA in the complementary direction. PCR takes advantage of this in order to focus on a particular portion of the DNA sequence.
PCR #
Suppose you find yourself in a situation where you want to examine a relatively small amount of DNA. This comes up fairly frequently, for instance in cases where you have an environmental sample or when you are looking for something—like the COVID virus—in a larger sample. In these cases, it's useful to amplify the DNA of interest so you have a larger amount for analysis. This is where the Polymerase Chain Reaction (PCR) comes in. PCR takes advantage of the same biological DNA replication mechanism I described above to amplify (make a lot of copies of) a DNA sequence of interest.
The basic idea is simple if you know the sequence of interest. (as with COVID, where we have the full sequence). You just synthesize primers that match both ends of the DNA sequence you want to replicate, with one matching the end in one direction and one matching the end in the other. When you mix them up with single-stranded DNA, the primers naturally hybridize (attach themselves) to the right places on the DNA strands. You then run the replication process with these primers.
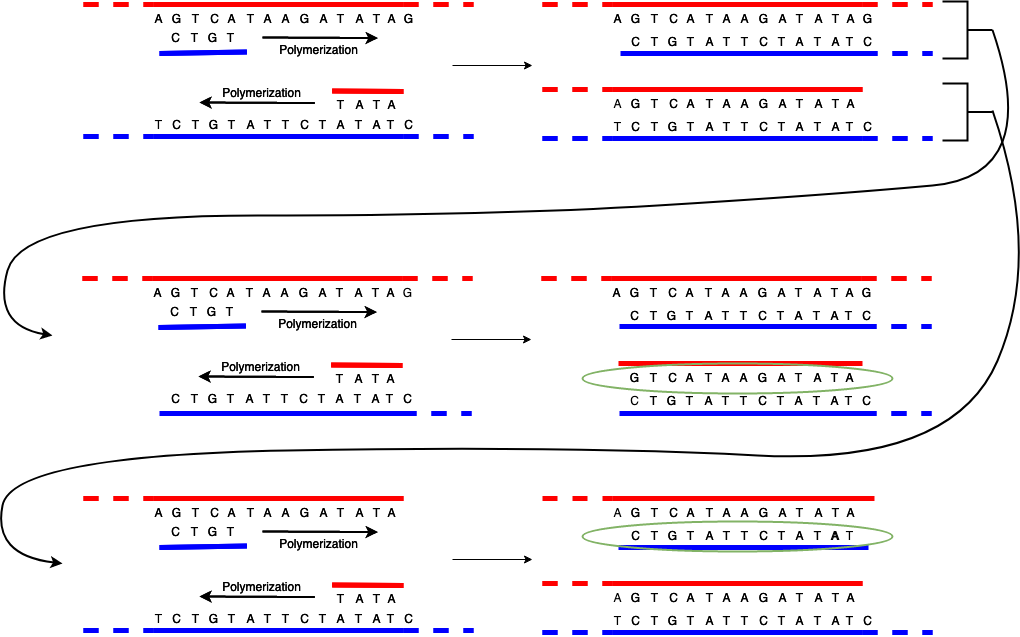
The first time you run the replication process, things are just as shown above: the strands separate and then the polymerase builds a partial replica of each original strand (on top of the original complementary strand) starting with the primer. At the end of this process you now have two paired DNA strands, as shown in the top part of the diagram below (which is just the same as the previous diagram.)
However, if you run the process again something interesting happens. As expected, each of the pairs unzips, leaving you with two original strands and then two partial replicas. The original strands replicate just as before and produce the same replicas as in the original process. However, when you build the complementary strand using the replicas from the first phase as the template they are built in the opposite direction from how that template strand was built. The result is that they start at the primer and stop when the strand ends, but because the strand already ended where the other primer was, the result is you get a strand that just consists of the region between the primers (inclusive). I've circled these strands in green so you can see them.
If you run the process over and over, what happens is that the original strands continue to make copies of partial strands and the partial (1st generation) and short (2nd and later generation) strands just make short strands. Each time you run the process you double the number of copies, so quite quickly you end up with a large copies of just the region of interest and a few copies of the rest of the DNA sample.
Partially Unknown Sequences #
Above, I assumed that you know the DNA sequence that you are interested in. This is certainly helpful, but it's not required. Actually, all you need to know is the sequence of the endpoints of the sequence of interest so you can make the primers. The replication process just depends on the primers binding to the relevant sections of DNA.[3] Once that happens, the polymerization process will work just fine with anything in between (in nature it obviously needs to work with basically any sequence). This is useful for a number of scenarios, such as:
-
When you want to sequence a specific piece of DNA, for instance to look for mutations or defects.
-
When you want to test for a DNA sequence that is subject to a lot of mutation, so you don't know exactly what's there (SARS-CoV-2, for instance, mutates quite rapidly).
In both situations, as long as you can find some surrounding regions that are highly conserved, you can make primers and replicate the region of interest.
PCR in Practice: Taq Polymerase #
Conceptually, then, PCR is simple: make the right primers, dump them into your sample, and then repeatedly run the replication cycle. But what does "run the replication cycle" actually mean? We need to unzip (melt) the DNA, then let polymerase make copies, and then repeat. But if we just dump some DNA, polymerase, primers, and bases into a tube, not much is going to happen because the DNA is already paired up, so we need something to kick off the process.
If you heat up the DNA to about 90°C, then it will melt, so you can heat it up, and then let it cool down a bit and it will replicate.[4] Unfortunately most polymerase enzymes are inactivated by being heated up, so if you just do this, you need to re-add polymerase every cycle, which is obviously a pain. However, the good news is that there is a polymerase enzyme (Taq polymerase) from a bacterium which lives in hot springs which can survive being heated to 90°C. This makes the problem much easier. You just need to mix up your DNA, primers, bases, and Taq polymerase in a tube and repeatedly heat it up and cool it down. You can buy a special machine called a thermal cycler that will do this automatically, and this is now standard lab equipment.[5]
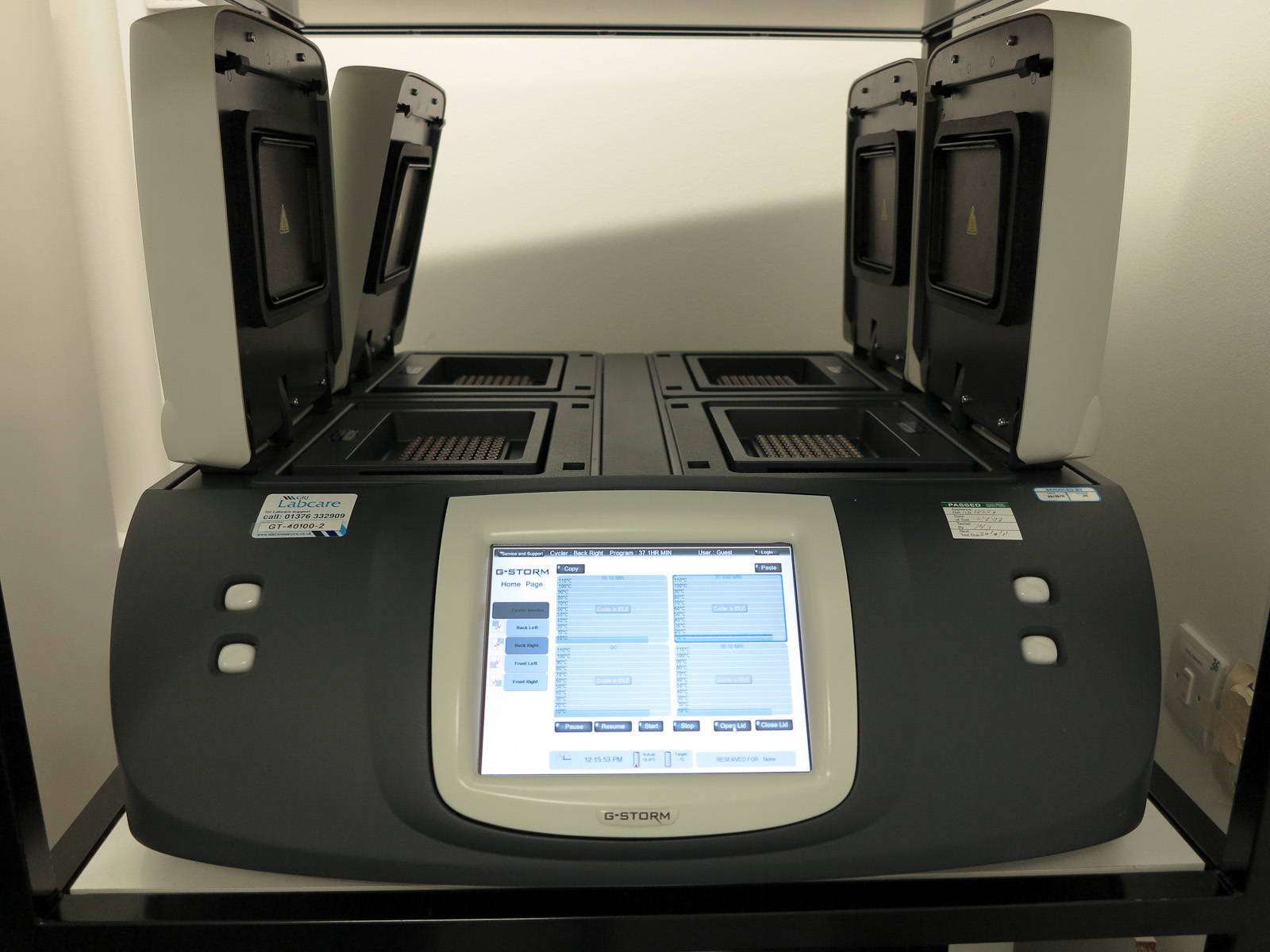
[From Rror via Wikipedia]
RNA #
OK, so this is all very useful, but what about if you want to amplify RNA? This is a particularly relevant application right now because SARS-CoV-2 (the virus that causes COVID-19) is an RNA virus (as is HIV). I'm not going to burden you with the details of RNA, except to say that (1) it's (usually) single-stranded rather than double-stranded and (2) it uses one different base[6] but is otherwise more or less is isomorphic to DNA. You can still PCR-amplify RNA by using an enzyme called reverse transcriptase that transcribes RNA into DNA, at which point PCR works as usual. This technique is called Reverse Transcriptase PCR. As far as I can tell, you basically can do RT-PCR by dumping reverse transcriptase into your sample and running things as usual.
PCR Testing #
OK, so I've told you how to amplify DNA sequences, which might be fine if you wanted to sequence them, but how do you use this to make a COVID test. The basic idea here is that you take a sample from the patient and look for COVID RNA in it (the same idea applies to HIV testing). But there's a missing step here because what I've described so far just replicates DNA, it doesn't measure it. I guess you could try to replicate things for a while and then maybe filter out the replicated strands and weigh them or something, but that would be a really tricky bit of analytical chemistry. Fortunately, there's a much cleverer approach, called Real-Time PCR or quantitative PCR (qPCR), which actually measures the replication process.
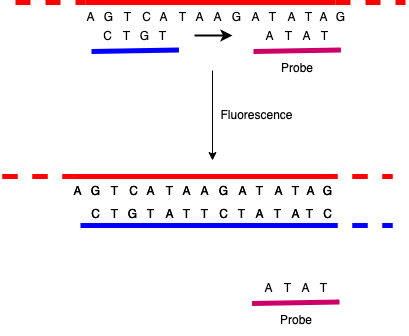
There are a number of ways to do this, but the basic trick is to measure replication via fluorescence. One version of this is to make a probe which is basically a DNA sequence that matches some sequence in the region of interest but also has some special chemistry that makes it fluoresce (glow) when it detaches from a strand of DNA. You then add that probe to the rest of the PCR mixture, where it adheres to the single strands much as the primers do.
When the PCR reaction runs, the polymerization process evicts the probe from the template strand, replacing it with the newly built complementary strand, causing it to fluoresce. You can then measure the light coming out as the reaction runs: the more replication that's happening—and hence the more DNA there is that matches the primers—the more light is emitted. Of course, because the PCR test inherently amplifies DNA sequences, if there is any significant amount of the target sequence, you'll eventually get some fluorescence, so what matters is the amount you see after a given number of PCR cycles. You'll sometimes hear the term cycle threshold (Ct) in connection which COVID tests. This is just the the number of cycles you had to run before you detected the virus. The more cycles, the less there was in the initial sample.
Final Thoughts #
If you're coming to this fresh, it's really hard to appreciate how revolutionary all this is, and how much it's come as the result of decades of hard work by thousands of talented scientists. Just what I've described here reflects at least three Nobel prizes (Crick, Watson, and Wilkins in 1962 for the discovery of the structure of DNA[7], Holey, Khorana, and Nirenberg for protein synthesis, Mullis for PCR), plus countless other contributions that didn't win the Nobel.
The result is an incredibly powerful set of analytic techniques—not just PCR, but fast sequencing, which I hope to talk about later—that have turned what used to be the effectively impossible problem of learning a given DNA sequence (the first viral DNA sequence was only performed back in 1984!) into what is today a routine task.
I'm really oversimplifying here. For instance, DNA can be methylated which affects how the DNA is interpreted without changing the sequence. ↩︎
In reality, of course, this process is messy and you get errors. There are also mechanisms to try to fix the errors, see proofreading). ↩︎
I think it's also possible to make multiple primers if there are variants, but I'm not a PCR expert. ↩︎
Your body, of course, does not heat up to 90°C, at least if you want to stay alive, but there are enzymes which will unzip the DNA at lower temperatures—2022-09-14. ↩︎
PCR was originally patented by Cetus and when I first saw PCR, the urban legend was that you could sell these machines without paying Cetus as long as you were careful to call them "thermal cyclers" rather than "PCR machines", even though as far as I know they were only used for PCR. ↩︎
Uracil rather than thymine ↩︎
See also Rosalind Franklin who did fundamental work here, but was widely overlooked for years. ↩︎